Studying the spatial distribution of proteomes in cells is crucial for understanding various molecular mechanisms in cell biology. In recent years, proximity chemical labeling technology, especially APEX (Engineered Ascorbate Peroxidase), has rapidly advanced. This method enables proximity labeling of proteins and RNA in specific spatial regions within living cells, avoiding false positives that often arise from traditional organelle separation techniques.
APEX technology uses peroxidase to catalyze the substrate biotin-phenol (BP), generating highly reactive free radicals that perform covalent addition reactions on nearby proteins and RNA, thus achieving spatial-specific labeling【1】. While APEX has been applied in various biological systems【2-5】, the large molecular size and low solubility of traditional BP substrates limit its use in organisms such as yeast and bacteria.
In response to these challenges, Researcher Zou Peng's group at Peking University has designed and synthesized a new generation of APEX probe substrate specifically for microbial systems—Alkyne-Phenol (Alk-Ph)【6】.
Compared to traditional BP substrates, Alk-Ph offers several key advantages:
- Smaller molecular size (MW: 217.27)
- Improved cell wall and membrane penetration
- Higher solubility in aqueous solutions
- Superior spatial specificity and an order of magnitude increase in labeling efficiency (Figure 1).

Figure 1 The higher efficiency of Alk-Ph labeling in yeast[6][6][6]. (A) Scheme of RNA labeling in yeast. (B) Real-time-PCR analysis of mitochondrial RNA enrichment elative to the cytoplasmic RNA markers, ACT1 and THD3.
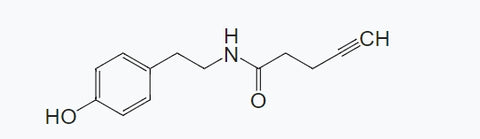
Alkyne-phenol, 95%
1694495-59-4
9186096
10 MG 50 MG
Transforming researcher Zou Peng's scientific research results, Bailingwei exclusively provides Alk-Ph, a protein and RNA proximity labeling probe used in microbial systems.

Researcher Zou Peng is currently at the Center for Synthetic and Functional Biomolecules at Peking University, where his work focuses on developing new chemical probe technologies. His innovative research aims to provide novel tools for neuroscience and related fields. By integrating protein engineering, molecular biology, and organic synthesis with advanced technologies such as optical microscopy, mass spectrometry, and high-throughput sequencing, his team can observe neuronal structure and activity. Through mathematical modeling, they perform quantitative data analysis, driving the study of nerve cells forward.
References
- Rhee, H.-W., Zou, P., Udeshi, N.D., Martell, J.D., Mootha, V.K., Carr, S.A., and Ting, A.Y. (2013). Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 339, 1328-1331.
- Hung, V., Lam, S.S., Udeshi, N.D., Svinkina, T., Guzman, G., Mootha, V.K., Carr, S.A., and Ting, A.Y. (2017). Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife 6, e24463.
- Hung, V., Udeshi, N.D., Lam, S.S., Loh, K.H., Cox, K.J., Pedram, K., Carr, S.A., and Ting, A.Y. (2016). Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc 11, 456-475.
- Hung, V., Zou, P., Rhee, H.-W., Udeshi, Namrata D., Cracan, V., Svinkina, T., Carr, Steven A., Mootha, Vamsi K., and Ting, Alice Y. (2014). Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol Cell 55, 332-341.
- Paek, J., Kalocsay, M., Staus, D.P., Wingler, L., Pascolutti, R., Paulo, J.A., Gygi, S.P., and Kruse, A.C. (2017). Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 169, 338-349.
- Li, Y., Tian, C., Liu, K., Zhou, Y., Yang, J., and Zou, P. (2020). A Clickable APEX Probe for Proximity-Dependent Proteomic Profiling in Yeast. Cell Chem Biol 27, 858-865.e858.
